Unit 23 - Spatio-temporal NDVI¶
Create a new mapset in oslo-region location, eg. sentinel-st-ndvi, see Work organization section.
Let’s download Sentinel L2A products for spring/summer 2017/2018. There are 4 products available as shown below. Note that in given period there are no Sentinel L2A products for 2018.
i.sentinel.download -l settings=sentinel.txt map=oslo area_relation=Contains \
start=2017-04-01 end=2017-10-01 producttype=S2MSI2A
2a894e37-1cf5-4bfc-ab42-9e32b99f423f 2017-05-23T10:40:31Z 1% S2MSI2Ap
71e0c5be-d008-4b71-a8f3-97f4c42ba09a 2017-05-06T10:50:31Z 2% S2MSI2Ap
74cf18cf-3cae-4d80-b1c8-9f2ee29972b4 2017-05-26T10:50:31Z 4% S2MSI2Ap
06724aba-9269-492c-a1d5-208a06c3f282 2017-07-05T10:50:31Z 27% S2MSI2Ap
Download selected Sentinel scenes:
i.sentinel.download settings=sentinel.txt map=oslo area_relation=Contains \
start=2017-05-01 end=2017-10-01 producttype=S2MSI2Ap output=geodata/sentinel/2017
Note
Pre-downloaded Sentinel scenes are available in sample
dataset geodata/sentinel/2017.
Data are imported by i.sentinel.import similarly as done in Unit 18 - Sentinel downloader. At fisrt check list of bands to be imported by -p flag. Since we are going to compute NDVI only 4th and 8th band are selected.
i.sentinel.import -p input=geodata/sentinel/2017 pattern="B0(4|8)_10m"
i.sentinel.import -l -c input=geodata/sentinel/2017 pattern="B0(4|8)_10m"
In next step a new space time raster dataset will be created and imported maps registered. Unfortunately there are no timestamps for imported maps required by t.register as shown in Unit 21. For this purpose a simple Python script has been designed.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 | #!/usr/bin/env python
#%module
#% description: Timestamps Sentinel bands from current mapset.
#%end
#%option G_OPT_F_OUTPUT
#%end
import sys
import os
from datetime import datetime, timedelta
import grass.script as gs
from grass.pygrass.gis import Mapset
def main():
mapset = Mapset()
mapset.current()
with open(options['output'], 'w') as fd:
for rast in mapset.glist('raster'):
items = rast.split('_')
d = datetime.strptime(items[2], '%Y%m%dT%H%M%S')
#fd.write("{0}|{1}{2}".format(rast, iso_date, os.linesep))
## workaround
dd = d + timedelta(seconds=1)
fd.write("{0}|{1}|{2}{3}".format(
rast,
d.strftime('%Y-%m-%d %H:%M:%S'),
dd.strftime('%Y-%m-%d %H:%M:%S'),
os.linesep))
return 0
if __name__ == "__main__":
options, flags = gs.parser()
sys.exit(main())
|
Timestamps can be easily determined from raster map name, for example L2A_T32UPB_20170517T102031_B04_10m raster map will be timestamped by 2017-05-17 10:20:31, see line 30.
Note
To avoid error in computation we also set up endtime timestamp as starttime+1sec, see 27.
Todo
This workaround should be avoided. Determine how?
Sample script to download: sentinel-timestamp.py
By running this script a timestamp file will be produced.
sentinel-timestamp.py output=sentinel-timestamps.txt
Example of created timestamps file show below.
L2A_T32VNM_20170523T104031_B04_10m|2017-05-23 10:40:31|2017-05-23 10:40:32
L2A_T32VNM_20170506T105031_B08_10m|2017-05-06 10:50:31|2017-05-06 10:50:32
L2A_T32VNM_20170526T105031_B04_10m|2017-05-26 10:50:31|2017-05-26 10:50:32
...
At this moment a new space time dataset can be created by t.create and all imported Sentinel bands registered by t.register.
t.create output=sentinel2017 title="Sentinel L2A 2017" desc="Oslo region NDVI computation"
t.register input=sentinel2017 file=sentinel-timestamps.txt
Let’s check basic metadata (t.info) and list of registered maps (t.rast.list).
t.info input=sentinel2017
...
| Start time:................. 2017-05-06 10:50:31
| End time:................... 2017-07-05 10:50:32
...
| Number of registered maps:.. 8
t.rast.list input=sentinel2017
name|mapset|start_time|end_time
L2A_T32VNM_20170506T105031_B04_10m|sentinel-st-ndvi|2017-05-06 10:50:31|2017-05-06 10:50:32
L2A_T32VNM_20170506T105031_B08_10m|sentinel-st-ndvi|2017-05-06 10:50:31|2017-05-06 10:50:32
L2A_T32VNM_20170523T104031_B04_10m|sentinel-st-ndvi|2017-05-23 10:40:31|2017-05-23 10:40:32
NDVI ST computation¶
For NDVI computation 4th and 8th bands are required (Unit 05 - Simple computation). Map algebra is performed in the case of spatio-temporal data by t.rast.mapcalc which requires data separated into spatio-temporal datasets (see example in Unit 22 - Spatio-temporal basic analysis). Such datasets can be prepared by t.rast.extract.
t.rast.extract input=sentinel2017 where="name like '%B04%'" output=b4
t.rast.extract input=sentinel2017 where="name like '%B08%'" output=b8
Let’s check content of the new datasets by t.rast.list.
t.rast.list input=b4
t.rast.list input=b8
Set computational region by g.region including mask for area of interest by r.mask.
g.region vector=oslo align=L2A_T32VNM_20170506T105031_B04_10m
r.mask vector=oslo
NDVI computation on spatio-temporal datasets can be performed in parallel (nproc).
t.rast.mapcalc input=b4,b8 output=ndvi expression="float(b8 - b4) / ( b8 + b4 )" \
basename=ndvi nproc=3
When computation is finished ndvi color table can be by t.rast.colors.
t.rast.colors input=ndvi color=ndvi
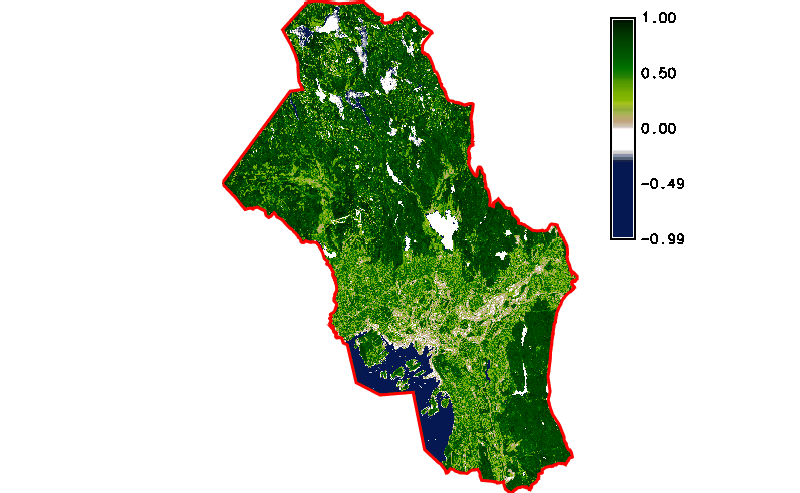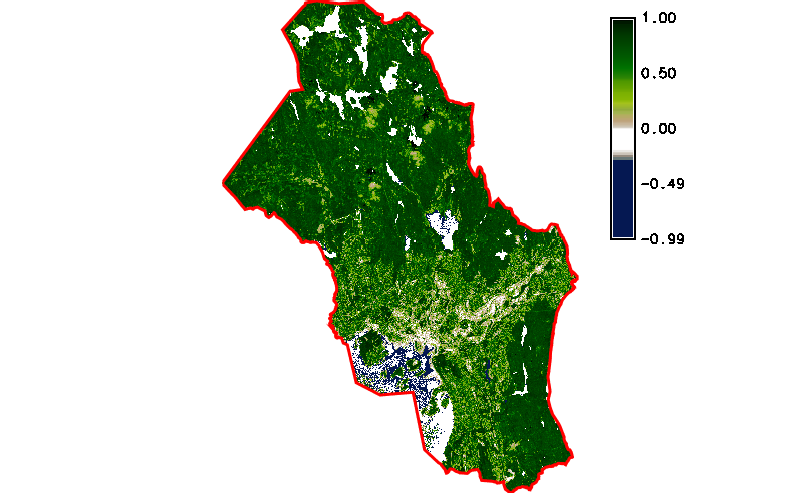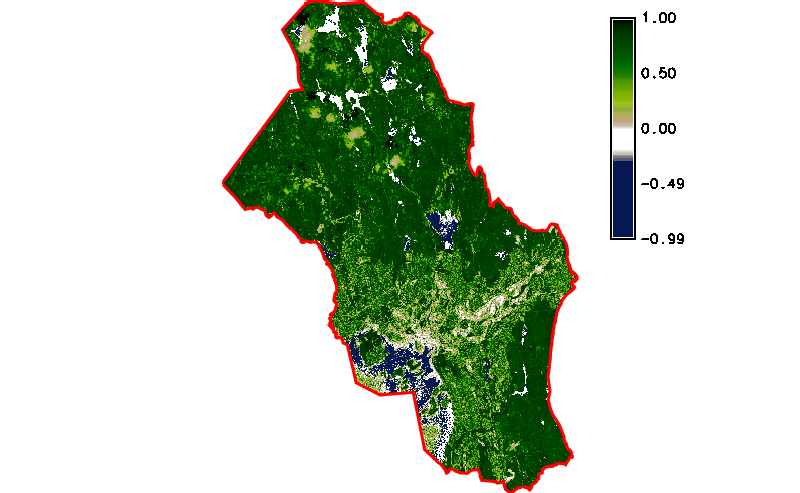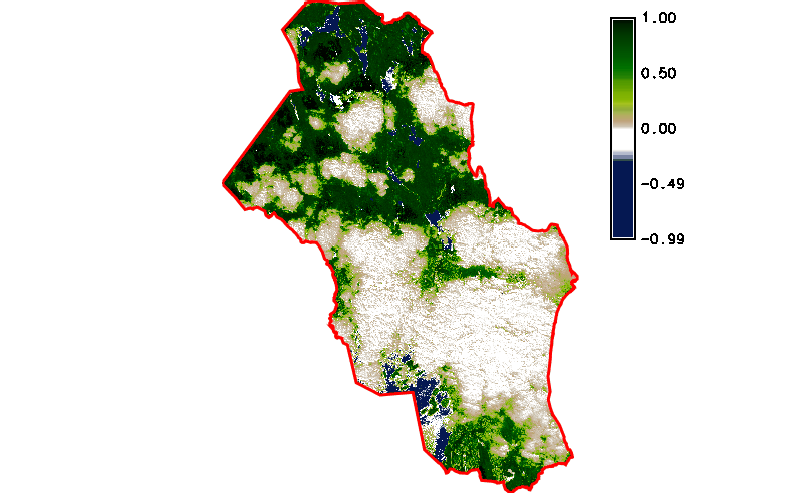
Fig. 121 Simple NDVI animation (no cloud mask applied) created by g.gui.animation.
Note
Load data as multiple raster maps instead of space time dataset. There is problem with sampling related to trick with endtime mentioned above.
Cloud mask¶
The result is not perfect enough. At least cloud mask should be
applied before NDVI computation. This operation can be easily solved
by applying new space time dataset containing computed raster masks. A
sample Python script has been designed for this purpose below. The
script also produces a timestamp file similarly to
sentinel-timestamp.py. Such mask can be created by
r.mask, see line 30. But in this case a mask
should be kept for further usage. Note that r.mask module
produces normal raster map with unique name MASK. To disable mask it
is enough to rename MASK map by g.rename, see line
43.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 | #!/usr/bin/env python
#%module
#% description: Creates raster mask maps based on clouds mask features.
#%end
#%option G_OPT_V_MAP
#% description: Name of AOI vector map
#%end
#%option G_OPT_F_OUTPUT
#%end
import sys
import os
from datetime import datetime, timedelta
import grass.script as gs
from grass.pygrass.gis import Mapset
from grass.pygrass.modules import Module
from grass.pygrass.vector import Vector
from grass.pygrass.utils import copy
def main():
mapset = Mapset()
mapset.current()
with open(options['output'], 'w') as fd:
for rast in mapset.glist('raster', pattern='*_B04_10m'):
items = rast.split('_')
d = datetime.strptime(items[2], '%Y%m%dT%H%M%S')
## workaround
dd = d + timedelta(seconds=1)
vect = '{}_{}_MSK_CLOUDS'.format(items[1], items[2])
mask_vect = '{}_{}'.format(vect, options['map'].split('@')[0])
if Vector(vect).exist():
Module('v.overlay', ainput=options['map'], binput=vect, operator='not',
output=mask_vect)
else:
copy(options['map'], mask_vect, 'vector')
Module('r.mask', vector=mask_vect, overwrite=True)
Module('g.remove', flags='f', type='vector', name=mask_vect)
Module('g.rename', raster=['MASK', mask_vect])
fd.write("{0}|{1}|{2}{3}".format(
mask_vect,
d.strftime('%Y-%m-%d %H:%M:%S'),
dd.strftime('%Y-%m-%d %H:%M:%S'),
os.linesep))
return 0
if __name__ == "__main__":
options, flags = gs.parser()
sys.exit(main())
|
Sample script to download: sentinel-cloud-mask.py
sentinel-cloud-mask.py map=oslo output=cloud-timestamps.txt
Now we can create a new space time dataset with raster cloud masks registered.
t.create output=cloud title="Sentinel L2A 2017 (cloud)" desc="Oslo region"
t.register input=cloud file=cloud-timestamps.txt
And apply modified expression for map algebra.
t.rast.mapcalc input=b4,b8,cloud output=ndvi_cloud \
expression="if(isnull(cloud), null(), float(b8 - b4) / ( b8 + b4 ))" \
basename=ndvi_cloud nproc=3
t.rast.colors input=ndvi_cloud color=ndvi
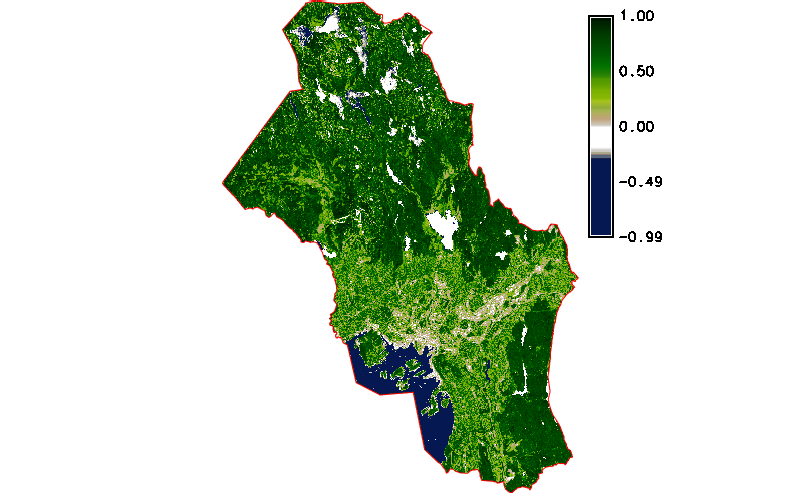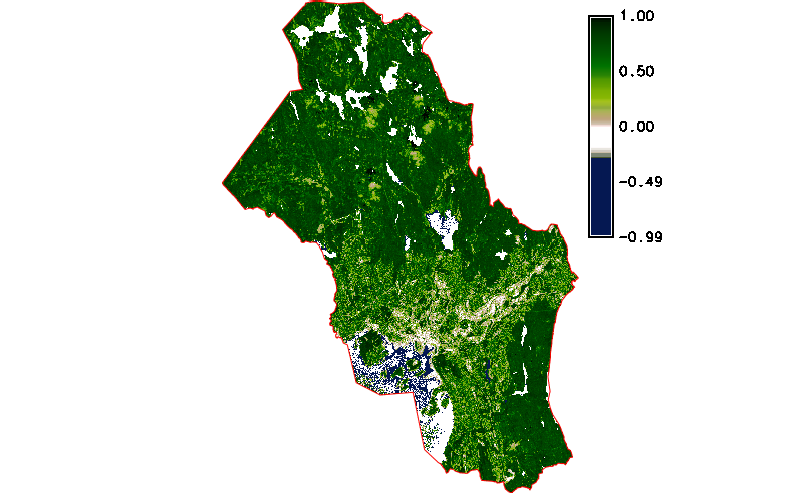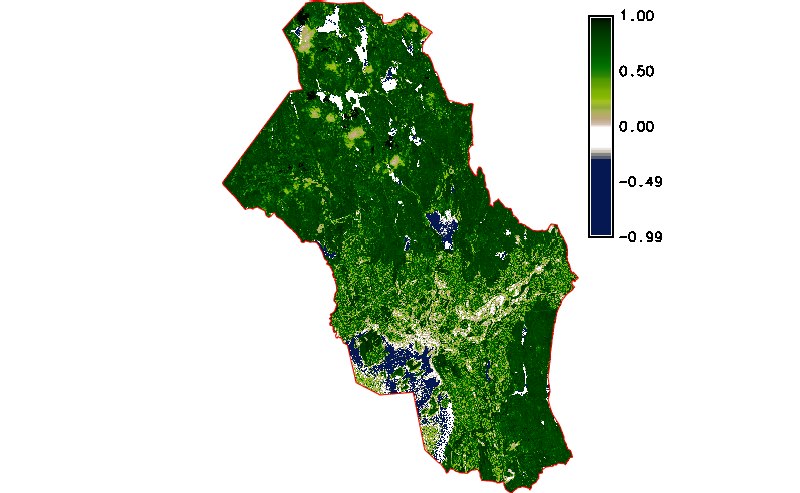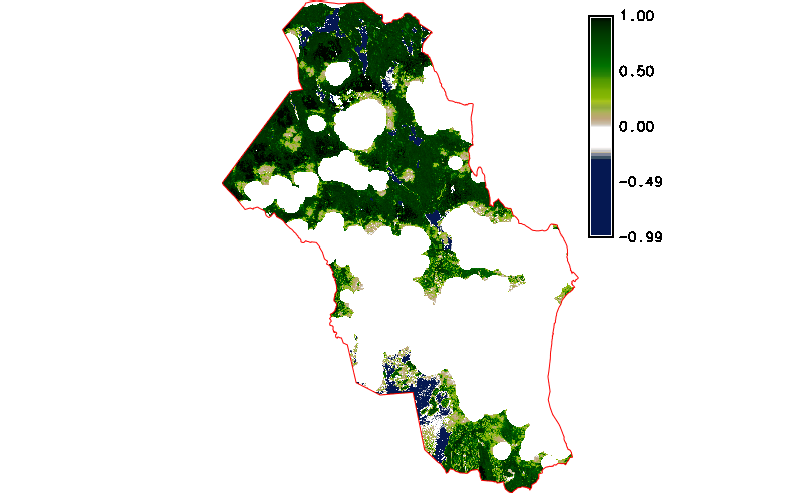
Fig. 122 Simple NDVI animation with cloud mask applied.