Unit 17 - DTM script parallelization¶
This unit is focused on computing parallelization. Sample script below produces seamless DTM (Digital Terrain Model, see Unit 16 - Lidar, DTM interpolation) from bunch of LAS/LAZ files. Computation will be split into tiles and performed in parallel.
DTM interpolation in parallel¶
User interface contains two major parameters, directory (line 6) for input directory with input LAS/LAZ files, and elevation (line 9) name for output elevation raster map mosaics. The resolution of output DTM is defined by resolution parameter (line 13). And finally number of processes running in parallel will be controlled by nproc (line 18) parameter.
A script consists of three main functions:
1. import_files() to import input LAS/LAZ files (line
33). Import process can be done in parallel by
ParallelModuleQueue from PyGRASS library (see
Unit 11 - PyGRASS scripting for PyGRASS intoruction), lines 37, 42, 57-58, 60.
2. create_dtm_tiles() to compute DTM per tile (line 67)
using v.surf.rst. DTM tiles need to be computed with a
reasonable overlap in order to create seamless mosaics, see
73-76. Tiles can be processed in parallel too, see
nproc option on line 82.
3. patch_tiles() to patch DTM tiles together by
r.series, see 86. From overlapping cell values is
computed an average value. This is main reason why r.patch
is not used here.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 | #!/usr/bin/env python
#%module
#% description: Creates DTM from input LAS tiles.
#%end
#%option G_OPT_M_DIR
#% required: yes
#%end
#%option G_OPT_R_ELEV
#% description: Name for output elevation raster map mosaics
#%end
#%option
#% key: resolution
#% description: Output resolution
#% type: double
#%end
#%option
#% key: nprocs
#% description: Number of processes per tile
#% answer: 1
#% type: integer
#%end
import os
import sys
import time
from copy import deepcopy
import grass.script as gs
from grass.pygrass.modules import Module, ParallelModuleQueue
def import_files(directory):
start = time.time()
# queue for parallel jobs
queue = ParallelModuleQueue(int(options['nprocs']))
import_module = Module('v.in.lidar',
flags='otb',
overwrite=True,
run_=False
)
maps = []
for f in os.listdir(directory):
if os.path.splitext(f)[1] != '.laz':
continue
fullname = os.path.join(directory, f)
basename = os.path.basename(f)
# '-' is not valid for vector map names
# vector map names cannot start with number
mapname = "las_{}".format(os.path.splitext(basename)[0].replace('-', '_'))
maps.append(mapname)
gs.message("Importing <{}>...".format(fullname))
import_task = deepcopy(import_module)
queue.put(import_task(input=fullname, output=mapname))
queue.wait()
if not maps:
gs.fatal("No input files found")
return maps
def create_dtm_tiles(maps, res, nprocs, offset_multiplier=10):
offset=res * offset_multiplier
for mapname in maps:
Module('g.region',
vector=mapname,
n='n+{}'.format(offset),
s='s-{}'.format(offset),
e='e+{}'.format(offset),
w='w-{}'.format(offset)
)
Module('v.surf.rst',
input=mapname,
elevation=mapname,
nprocs=nprocs,
overwrite=True
)
def patch_tiles(maps, output, resolution):
gs.message("Patching tiles <{}>...".format(','.join(maps)))
Module('g.region', raster=maps, res=resolution)
Module('r.series', input=maps, output=output, method='average', overwrite=True)
Module('r.colors', map=output, color='elevation')
def main():
start = time.time()
maps = import_files(options['input'])
create_dtm_tiles(maps,
float(options['resolution']),
int(options['nprocs'])
)
patch_tiles(maps,
options['elevation'],
options['resolution']
)
gs.message("Done in {:.0f} min".format((time.time() - start) / 60.))
return 0
if __name__ == "__main__":
options, flags = gs.parser()
sys.exit(main())
|
Sample script to download: create-dtm.py
Note
The script is taking a long time with all the tiles from
/home/geodata/lidar/laz directory. Choose few tiles
for testing.
Create a new directory eg. /tmp/lidar and link some (2
or 3) of the LAZ files with
ln -s /home/geodata/lidar/laz/32-1-514-136-15.laz /tmp/lidar/
Use /tmp/lidar as input in the create-dtm.py script
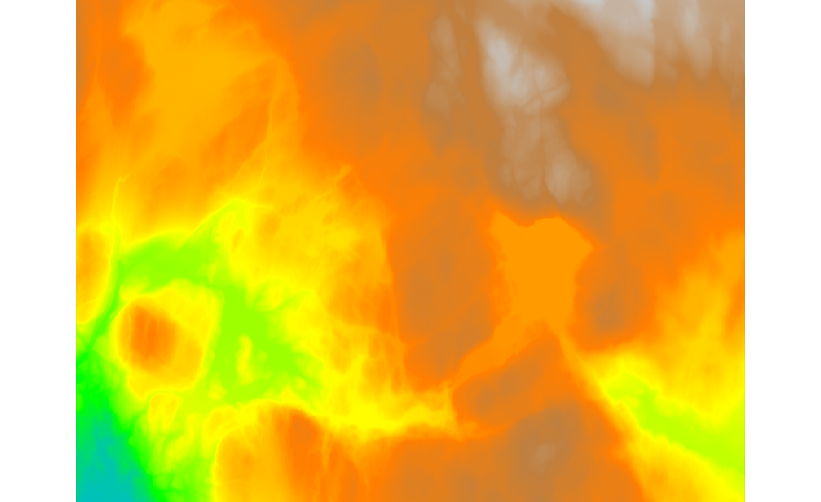
Fig. 100 DTM created from all available tiles.
DTM comparision¶
In this session we are going to calculate the Canopy Height Model (CHM), it is the difference between interpolated DSM and imported EUDEM DTM
The CHM is computed using r.mapcalc, executing the difference between DSM and DTM
r.mapcalc expression="chm = DTM_laz - dem"
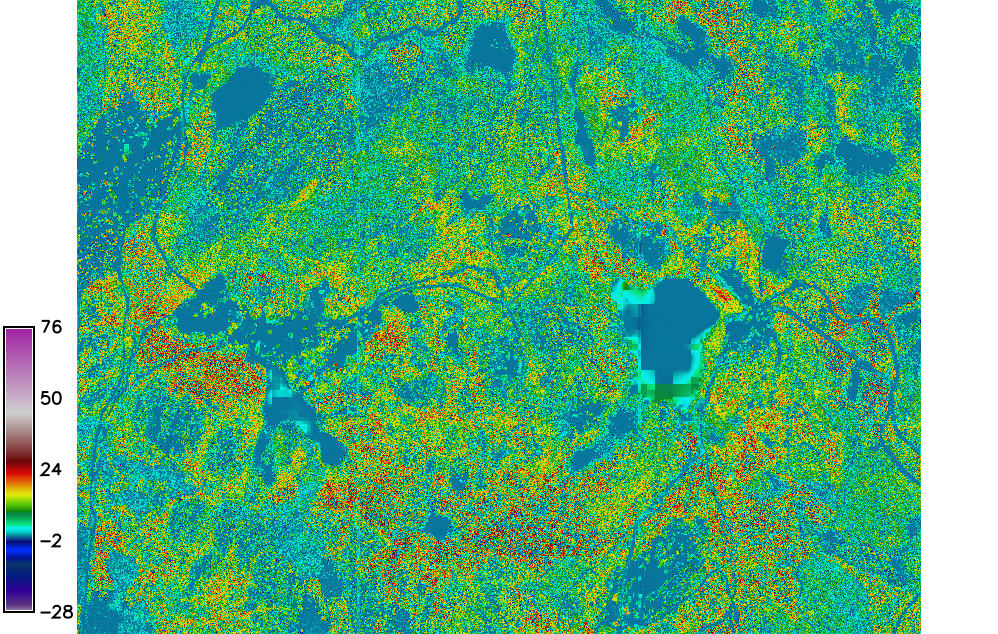
Fig. 101 The CHM map.
Tip
To have better CHM it is possible to download DTM files from hoydedata.no. You can import all TIF files using checking Directory as Source type and selecting the directory where the TIF files are.
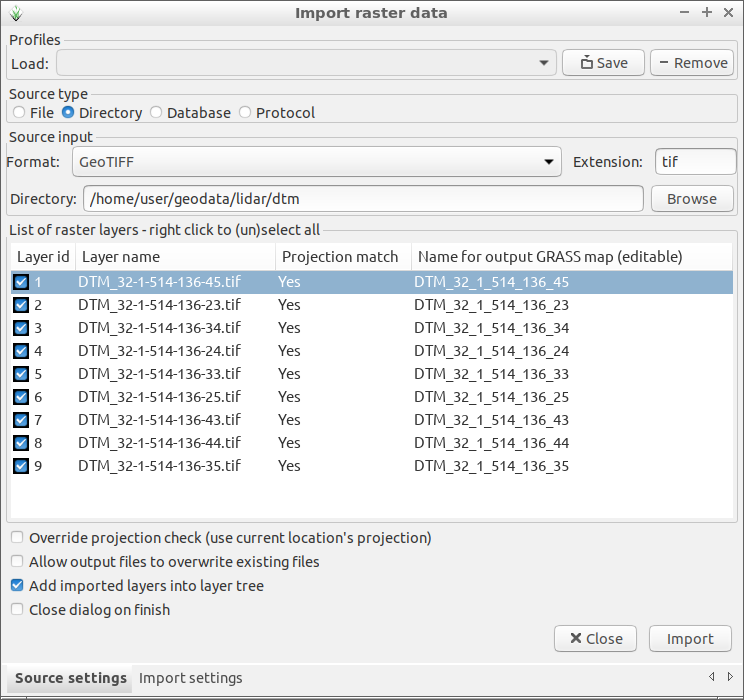
Fig. 102 Import all DTM TIF files from for exampe geodata/lidar/dtm/
directory.
Once DTM tiles are imported you can patch them using r.series and set the color to elevation by r.colors.
r.series input=`g.list type=raster pattern=DTM_* sep=','` output=DTM_patch
r.colors map=DTM_patch color=elevation

Fig. 103 Patched DTM.
Now it is possible to run the previous r.mapcalc command changing dem with DTM_patch